- Facility
Epitranscriptomics & Sequencing (EpiRNA-Seq)
- Next-Generation Sequencing, RNA, RNA modifications

The Epitranscriptomics and Sequencing (EpiRNA-Seq) Core Facility is built on the expertise of the UMR7365 IMoPA in the field of RNA molecular biology to promote its main collaborative services.
Located on the Brabois-Santé campus of the University of Lorraine in Nancy, it provides to the academic and industrial scientific community high-tech resources (Illumina next generation sequencers). It is part of the European COST EPITRAN network and regularly organises training activities.
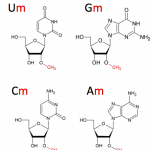
The core facility has recognized technical and methodological know-how to contribute to various projects in Epitranscriptomics (RiboMethSeq, AlkAniline-Seq, HydraPsiSeq) to address various scientific questions such as dynamics of post-transcriptional modifications in RNAs.
The EpiRNA-Seq Core Facility operates mainly in collaborative mode (involvement of members of the core facility in the steps of your sequencing project from the conception of the experimental design to the statistical analysis of the data) but the delivery mode (mainly sequencing of libraries prepared by you without involvement of the platform) is also possible under certain conditions. The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academics and industrialists).
The EpiRNA-Seq Core Facility is open to the academic and industrial scientific community at the local, national and international level.
The team is at your disposal to help you by bringing its technical and methodological know-how and to follow you from A to Z in your sequencing projects.
The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academic and industrial).
Any new collaborator must first contact the head of the core facility using the "Request form" to define the best strategy to adopt for conducting his study, and obtain a quote. Any user will agree to accept the "terms and conditions". The guidelines to prepare and send samples to the Core Facility are given in "Sample sheet".
Marcel V, Kielbassa J, Marchand V, Natchiar SK, Paraqindes H, Nguyen Van Long F, Ayadi L, Bourguignon-Igel V, Lo Monaco P, Montchiet D, Scott V, Tonon L, Bray SE, Diot A, Jordan LB, Thompson AM, Bourdon JC, Dubois T, André F, Catez F, Puisieux A, Motorin Y, Klaholz BP, Viari A, Diaz JJ. Ribosomal RNA 2′O-methylation as a novel layer of inter-tumour heterogeneity in breast cancer. NAR Cancer. 2020 Dec 22:zcaa036.
 10.1093/narcan/zcaa036 ,
10.1093/narcan/zcaa036 ,  inserm-03103238
inserm-03103238 Werner S, Galliot A, Pichot F, Kemmer T, Marchand V, Sednev MV, Lence T, Roignant JY, König J, Höbartner C, Motorin Y, Hildebrandt A, Helm M. NOseq: amplicon sequencing evaluation method for RNA m6A sites after chemical deamination. Nucleic Acids Res. 2020 Dec 11:gkaa1173.
 10.1093/nar/gkaa1173 ,
10.1093/nar/gkaa1173 ,  33313868 ,
33313868 ,  HAL-03093648
HAL-03093648 Galvanin A, Vogt LM, Grober A, Freund I, Ayadi L, Bourguignon-Igel V, Bessler L, Jacob D, Eigenbrod T, Marchand V, Dalpke A, Helm M, Motorin Y. Bacterial tRNA 2'-O-methylation is dynamically regulated under stress conditions and modulates innate immune response. Nucleic Acids Res. 2020 Dec 4:gkaa1123.
 10.1093/nar/gkaa1123 ,
10.1093/nar/gkaa1123 ,  33275131 ,
33275131 ,  HAL-03071703
HAL-03071703 Marchand V, Pichot F, Neybecker P, Ayadi L, Bourguignon-Igel V, Wacheul L, Lafontaine DLJ, Pinzano A, Helm M, Motorin Y. HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 2020 Sep 25 :gkaa769.
 10.1093/nar/gkaa769 ,
10.1093/nar/gkaa769 ,  32976574 ,
32976574 ,  HAL-02957799
HAL-02957799 Kristen M, Plehn J, Marchand V, Friedland K, Motorin Y, Helm M, Werner S. Manganese Ions Individually Alter the Reverse Transcription Signature of Modified Ribonucleosides. Genes (Basel). 2020 Aug 18;11(8):E950.
 10.3390/genes11080950 ,
10.3390/genes11080950 ,  32824672 ,
32824672 ,  HAL-02929785
HAL-02929785 Human GTPBP5 (MTG2) fuels mitoribosome large subunit maturation by facilitating 16S rRNA methylation. Nucleic Acids Res. 2020 Jul 11 : gkaa592.
 10.1093/nar/gkaa592 ,
10.1093/nar/gkaa592 ,  32652011
32652011 De Crécy-Lagard V, Ross RL, Jaroch M, Marchand V, Eisenhart C, Brégeon D, Motorin Y, Limbach PA. Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis Sp Subtilis Strain 168. Biomolecules. 2020 Jun 30;10(7):E977.
 10.3390/biom10070977 ,
10.3390/biom10070977 ,  32629984 ,
32629984 ,  HAL-02915956
HAL-02915956 Trzaska C, Amand S, Bailly C, Leroy C, Marchand V, Duvernois-Berthet E, Saliou JM, Benhabiles H, Werkmeister E, Chassat T, Guilbert R, Hannebique D, Mouray A, Copin MC, Moreau PA, Adriaenssens E, Kulozik A, Westhof E, Tulasne D, Motorin Y, Rebuffat S, Lejeune F. 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat Commun. 2020 Mar 20.
 10.1038/s41467-020-15140-z ,
10.1038/s41467-020-15140-z ,  32198346 ,
32198346 ,  HAL-02515244
HAL-02515244 Pichot F, Marchand V, Ayadi L, Bourguignon-Igel V, Helm M, Motorin Y. Holistic Optimization of Bioinformatic Analysis Pipeline for Detection and Quantification of 2'-O-Methylations in RNA by RiboMethSeq. Front Genet. 2020 Feb 13.
 10.3389/fgene.2020.00038 ,
10.3389/fgene.2020.00038 ,  32117451 ,
32117451 ,  HAL-02498335
HAL-02498335 Werner S, Schmidt L, Marchand V, Kemmer T, Falschlunger C, Sednev MV, Bec G, Ennifar E, Höbartner C, Micura R, Motorin Y, Hildebrandt A, Helm M. Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res. 2020 Feb 25.
 10.1093/nar/gkaa113 ,
10.1093/nar/gkaa113 ,  32095818 ,
32095818 ,  HAL-02492494
HAL-02492494

Formation CNRS Entreprises
( -> )
The EpiRNA-Seq Core Facility organizes the following training school (CNRS Formation Entreprises) : Epitranscriptomics: mapping and analysis of RNA modifications by Next-Generation Sequencing. This training school will be held from 17st to 19st May 2021 on the Core Facility in Nancy. For more details : https://cnrsformation.cnrs.fr/epitranscriptomics-mapping-and-analysis-of-rna-modifications-by-next-generation-sequencing?axe=140

Epitranscriptomics training school (COST Epitran)
( -> )
The NGS Core Facility in collaboration with Pr Iouri Motorine (UMR7365 IMoPA) is organizing a training school on Epitranscriptomics with the support of COST Epitran from 11-14 february 2019 in Nancy (France). This training is dedicated to young phD students or postdocs. The session in 2019 is closed. The next session will take place in February 2020, please send an email to the head of EpiRNA-Seq core facility. For more information : /sites/default/files/users/Projets/Diapositive1.png