Facility
Epitranscriptomics & Sequencing (EpiRNA-Seq)
Epitranscriptome / Analyses of RNA modifications
Actually, the EpiRNA-Seq core facility proposes the following epitranscriptomic analyses :
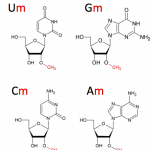
- Analysis of rRNA/tRNA and mRNA 2’-O-methylations by RiboMethSeq (Marchand et al., 2016).
This method was developed by the group of Y. Motorin about 3 years ago. It allows the mapping at single nucleotide resolution and the quantification of 2'-O-methylations present in rRNA or tRNA with 10 ng of RNA as input material.
The EpiRNA-Seq core facility was involved in major scientific projects such as :
- Analysis of 2'-O-methylation deregulation in rRNA in cancer linked to Fibrillarin and snoRNA decrease (Erales, Marchand et al., 2017 and Sharma, Marchand et al., 2017),
- Analysis of rRNA/tRNA modification profiling in bacterial or eukaryotic stress and development. In particular, 2’-O-Me in tRNA is heavily involved in innate immunity (Gehrig et al., 2012) and, as we recently demonstrated, in HIV-1 virus recognition in infected cell (Ringeard, Marchand et al., 2019).
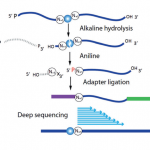
- Analysis of m7G/m3C/D modifications in RNAs by AlkAniline-Seq (Marchand et al., 2018).
This method was recently developed by the core facility (Marchand et al., 2018), but represents an important perspective for extension of RNA modification analysis. AlkAniline-Seq is dedicated to the study of internal m7G/m3C/D residues in all kind of RNAs starting with 150-300 ng of total RNA as input material.
Several projects require the precise mapping of those modifications in different RNAs, notably in relation to antibiotic resistance in bacteria, as well as in mitochondrial diseases related to tRNA deficiency.
- Analysis of pseudouridines in RNAs by HydraPsi-Seq (Marchand et al., 2020).
HydraPsi-Seq is a robust method for systematic mapping at single nucleotide resolution and accurate quantification of pseudouridines residues in RNA. An interesting point is that it requires minute amounts (10-50 ng) as input material making it compatible with high-throughput profiling of diverse biological and clinical samples.
- Request form160.17 KB
- Terms and conditions215.97 KB
These analyses are carried out in collaborative mode. If you are interested, please contact us using the "Request form" to define the best strategy to adopt for conducting your study, and obtain a quote. Any user will agree to accept the terms and conditions.
Marchand V, Pichot F, Neybecker P, Ayadi L, Bourguignon-Igel V, Wacheul L, Lafontaine DLJ, Pinzano A, Helm M, Motorin Y. HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 2020 Sep 25 :gkaa769.
 10.1093/nar/gkaa769 ,
10.1093/nar/gkaa769 ,  32976574 ,
32976574 ,  HAL-02957799
HAL-02957799 Marchand V, Ayadi L, Ernst FGM, Hertler J, Bourguignon-Igel V, Galvanin A, Kotter A, Helm M, Lafontaine DLJ, Motorin Y. AlkAniline-Seq: profiling of m7G and m3C RNA modifications at single nucleotide resolution. Angew Chem Int Ed Engl.2018 Oct 29.
 10.1002/anie.201810946 ,
10.1002/anie.201810946 ,  30370969 ,
30370969 ,  HAL-01928435
HAL-01928435 Marchand V, Ayadi L, El Hajj A, Blanloeil-Oillo F, Helm M, Motorin Y. High-Throughput Mapping of 2'-O-Me Residues in RNA Using Next-Generation Sequencing (Illumina RiboMethSeq Protocol). Methods Mol Biol. 2017 ; 1562:171-187.
 10.1007/978-1-4939-6807-7_12 ,
10.1007/978-1-4939-6807-7_12 ,  28349461 ,
28349461 ,  HAL-01799289
HAL-01799289 Marchand V, Pichot F, Thüring K, Ayadi L, Freund I, Dalpke A, Helm M, Motorin Y. Next-Generation Sequencing-Based RiboMethSeq Protocol for Analysis of tRNA 2'-O-Methylation. Biomolecules. 2017 Feb 9 ; 7(1).
 10.3390/biom7010013 ,
10.3390/biom7010013 ,  28208788 ,
28208788 ,  HAL-01799272
HAL-01799272 Sharma S, Marchand V, Motorin Y, Lafontaine DLJ. Identification of sites of 2'-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci Rep. 2017 Sep 13 ; 7(1):11490.
 10.1038/s41598-017-09734-9 ,
10.1038/s41598-017-09734-9 ,  28904332 ,
28904332 ,  HAL-01799319
HAL-01799319 Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, Garcia M, Laforêts F, Marcel V, Baudin-Baillieu A, Bertin P, Couté Y, Adrait A, Meyer M, Therizols G, Yusupov M, Namy O, Ohlmann T, Motorin Y, Catez F, Diaz JJ. Evidence for rRNA 2'-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci U S A. 2017 Dec 5 ; 114(49):12934-12939.
 10.1073/pnas.1707674114 ,
10.1073/pnas.1707674114 ,  29158377 ,
29158377 ,  HAL-01866982
HAL-01866982 Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. FTSJ3 is an RNA 2'-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019 Jan 9.
 10.1038/s41586-018-0841-4 ,
10.1038/s41586-018-0841-4 ,  30626973 ,
30626973 ,  HAL-01981983
HAL-01981983 Galvanin A, Ayadi L, Helm M, Motorin Y, Marchand V. Mapping and Quantification of tRNA 2'-O-Methylation by RiboMethSeq. Methods Mol Biol. 2019 ; 1870:273-295.
 10.1007/978-1-4939-8808-2_21 ,
10.1007/978-1-4939-8808-2_21 ,  30539563 ,
30539563 ,  HAL-01957102
HAL-01957102 Ayadi L, Motorin Y, Marchand V. Quantification of 2'-O-Me Residues in RNA Using Next-Generation Sequencing (Illumina RiboMethSeq Protocol). Methods Mol Biol. 2018 ; 1649:29-48.
 10.1007/978-1-4939-7213-5_2 ,
10.1007/978-1-4939-7213-5_2 ,  29130188 ,
29130188 ,  HAL-01661952
HAL-01661952