- Facility
Epitranscriptomics & Sequencing (EpiRNA-Seq)
- Next-Generation Sequencing, RNA, RNA modifications

The Epitranscriptomics and Sequencing (EpiRNA-Seq) Core Facility is built on the expertise of the UMR7365 IMoPA in the field of RNA molecular biology to promote its main collaborative services.
Located on the Brabois-Santé campus of the University of Lorraine in Nancy, it provides to the academic and industrial scientific community high-tech resources (Illumina next generation sequencers). It is part of the European COST EPITRAN network and regularly organises training activities.
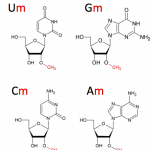
The core facility has recognized technical and methodological know-how to contribute to various projects in Epitranscriptomics (RiboMethSeq, AlkAniline-Seq, HydraPsiSeq) to address various scientific questions such as dynamics of post-transcriptional modifications in RNAs.
The EpiRNA-Seq Core Facility operates mainly in collaborative mode (involvement of members of the core facility in the steps of your sequencing project from the conception of the experimental design to the statistical analysis of the data) but the delivery mode (mainly sequencing of libraries prepared by you without involvement of the platform) is also possible under certain conditions. The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academics and industrialists).
The EpiRNA-Seq Core Facility is open to the academic and industrial scientific community at the local, national and international level.
The team is at your disposal to help you by bringing its technical and methodological know-how and to follow you from A to Z in your sequencing projects.
The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academic and industrial).
Any new collaborator must first contact the head of the core facility using the "Request form" to define the best strategy to adopt for conducting his study, and obtain a quote. Any user will agree to accept the "terms and conditions". The guidelines to prepare and send samples to the Core Facility are given in "Sample sheet".
- Groza P, Kumari K, Esteva-Socias M, Schott J, Bhattarai DP, Sajkowska JJ, Dasgupta R, Peula C, Destefanis E, Williams C, Marchand V, Wikström P, Wiberg R, Campos AB, Gilthorpe JD, Pop B, Mateus A, Motorin Y, Dassi E, Petzold K, Tuorto F, Aguilo F. Fibrillarin-dependent 2'-O-methylation modulates RPS28 ribosome incorporation and oncogenic translation. Cancer Lett. 2025 Nov 17:218124.
 10.1016/j.canlet.2025.218124 ,
10.1016/j.canlet.2025.218124 ,  41260515 ,
41260515 ,  HAL-05376009
HAL-05376009 - Lebret-Kogey V, Ayadi L, Maenner S, Motorin Y, Behm-Ansmant I, Aigueperse C. Quantitative detection of 2'-O-methylated residues in non-coding RNAs using DNAzymes and quantitative RT-PCR. Biochimie. 2025 Oct;237:26-39.
 10.1016/j.biochi.2025.07.006 ,
10.1016/j.biochi.2025.07.006 ,  40633801 ,
40633801 ,  HAL-05162861
HAL-05162861 - Rabany O, Ben Dror S, Arafat M, Aharoni Levitanus H, Halperin Y, Marchand V, Romanovski N, Ussishkin N, Livneh Golany M, Reches A, Wexler J, Mayorek N, Monderer-Rothkoff G, Shifman S, Mâmmer Bouhou W, VanInsberghe M, Pauli C, Müller-Tidow C, Karmi O, Livneh Y, van Oudenaarden A, Motorin Y, Nachmani D. Dynamic rRNA Methylation Regulates Translation in the Hematopoietic System and is Essential for Stem Cell Fitness. Blood. 2025 Nov 5:blood.2024028300.
 10.1182/blood.2024028300 ,
10.1182/blood.2024028300 ,  41191530 ,
41191530 ,  HAL-05353198
HAL-05353198 - Bacci L, Pollutri D, Ripa IJ, D'Andrea M, Marchand V, Motorin Y, Hesse AM, Couté Y, Filipek K, Penzo M. Ribosomal protein L5 (RPL5/uL18) I60V mutation is associated to increased translation and modulates drug sensitivity in T-cell acute lymphoblastic leukemia cells. Biochem Pharmacol. 2025 Oct 31;243(Pt 1):117497.
 10.1016/j.bcp.2025.117497 ,
10.1016/j.bcp.2025.117497 ,  41177179 ,
41177179 ,  HAL-05353080
HAL-05353080 - Hardy L, Marchand V, Bourguignon V, Thuillier Q, Dias C, Krin E, Fruchard L, Yaacov DB, Mazel D, Motorin Y, Baharoglu Z. The tRNA epitranscriptomic landscape and RNA modification enzymes in Vibrio cholerae. PLoS Genet. 2025 Oct 31;21(10):e1011937.
 10.1371/journal.pgen.1011937 ,
10.1371/journal.pgen.1011937 ,  41171884 ,
41171884 ,  Pasteur-05347080
Pasteur-05347080 - International Human RNome Project Consortium. Unlocking the regulatory code of RNA: launching the Human RNome Project. Genome Biol. 2025 Oct 24;26(1):367.
 10.1186/s13059-025-03824-y ,
10.1186/s13059-025-03824-y ,  41137159
41137159 - Belukhina S, Saudemont B, Depardieu F, Lorthios T, P Maviza T, Livenskyi A, Serebryakova M, Aleksandrova M, Ukholkina E, Burmistrova N, Sergiev P, Libiad M, Dubrac S, Barras F, Motorin Y, Marchand V, Hagelueken G, Isaev A, Bikard D, Rouillon C. Specificity and mechanism of tRNA cleavage by the AriB Toprim nuclease of the PARIS bacterial immune system. Philos Trans R Soc Lond B Biol Sci. 2025 Sep 4;380(1934):20240074.
 10.1098/rstb.2024.0074 ,
10.1098/rstb.2024.0074 ,  40904115 ,
40904115 ,  HAL-05353021
HAL-05353021 - Koenig L, Guggenberger V, Eleftheriou K, Pinter Z, Marotto A, Kreutz CR, Wossidlo M, Marchand V, Motorin Y, Schaefer MR. Copy Number Determination of Sperm-Borne Small RNAs Implied in the Intergenerational Inheritance of Metabolic Syndromes. RNA. 2025 May 28:rna.080480.125.
 10.1261/rna.080480.125 ,
10.1261/rna.080480.125 ,  40436631 ,
40436631 ,  HAL-05106950
HAL-05106950 - Leroy E, Challal D, Pelletier S, Goncalves C, Menant A, Marchand V, Jaszczyszyn Y, van Dijk E, Naquin D, Andreani J, Motorin Y, Palancade B, Rougemaille M. A bifunctional snoRNA with separable activities in guiding rRNA 2'-O-methylation and scaffolding gametogenesis effectors. Nat Commun. 2025 Apr 5;16(1):3250.
 10.1038/s41467-025-58664-y ,
10.1038/s41467-025-58664-y ,  40185772 ,
40185772 ,  HAL-05106967
HAL-05106967 - Morin C, Paraqindes H, Van Long FN, Isaac C, Thomas E, Pedri D, Pulido-Vicuna CA, Morel AP, Marchand V, Motorin Y, Carrere M, Auclair J, Attignon V, Pommier RM, Ruiz E, Bourdelais F, Catez F, Durand S, Ferrari A, Viari A, Marine JC, Puisieux A, Diaz JJ, Moyret-Lalle C, Marcel V. Specific modulation of 28S_Um2402 rRNA 2'-O-ribose methylation as a novel epitranscriptomic marker of ZEB1-induced epithelial-mesenchymal transition in different mammary cell contexts. NAR Cancer. 2025 Jan 28;7(1):zcaf001.
 10.1093/narcan/zcaf001 ,
10.1093/narcan/zcaf001 ,  39877292 ,
39877292 ,  HAL-05021643
HAL-05021643

Formation CNRS Entreprises
( -> )
The EpiRNA-Seq Core Facility organizes the following training school (CNRS Formation Entreprises) : Epitranscriptomics: mapping and analysis of RNA modifications by Next-Generation Sequencing. This training school will be held from 17st to 19st May 2021 on the Core Facility in Nancy. For more details : https://cnrsformation.cnrs.fr/epitranscriptomics-mapping-and-analysis-of-rna-modifications-by-next-generation-sequencing?axe=140

Epitranscriptomics training school (COST Epitran)
( -> )
The NGS Core Facility in collaboration with Pr Iouri Motorine (UMR7365 IMoPA) is organizing a training school on Epitranscriptomics with the support of COST Epitran from 11-14 february 2019 in Nancy (France). This training is dedicated to young phD students or postdocs. The session in 2019 is closed. The next session will take place in February 2020, please send an email to the head of EpiRNA-Seq core facility. For more information : /sites/default/files/users/Projets/Diapositive1.png