- Facility
Epitranscriptomics & Sequencing (EpiRNA-Seq)
- Next-Generation Sequencing, RNA, RNA modifications

The Epitranscriptomics and Sequencing (EpiRNA-Seq) Core Facility is built on the expertise of the UMR7365 IMoPA in the field of RNA molecular biology to promote its main collaborative services.
Located on the Brabois-Santé campus of the University of Lorraine in Nancy, it provides to the academic and industrial scientific community high-tech resources (Illumina next generation sequencers). It is part of the European COST EPITRAN network and regularly organises training activities.
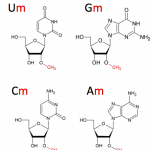
The core facility has recognized technical and methodological know-how to contribute to various projects in Epitranscriptomics (RiboMethSeq, AlkAniline-Seq, HydraPsiSeq) to address various scientific questions such as dynamics of post-transcriptional modifications in RNAs.
The EpiRNA-Seq Core Facility operates mainly in collaborative mode (involvement of members of the core facility in the steps of your sequencing project from the conception of the experimental design to the statistical analysis of the data) but the delivery mode (mainly sequencing of libraries prepared by you without involvement of the platform) is also possible under certain conditions. The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academics and industrialists).
The EpiRNA-Seq Core Facility is open to the academic and industrial scientific community at the local, national and international level.
The team is at your disposal to help you by bringing its technical and methodological know-how and to follow you from A to Z in your sequencing projects.
The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academic and industrial).
Any new collaborator must first contact the head of the core facility using the "Request form" to define the best strategy to adopt for conducting his study, and obtain a quote. Any user will agree to accept the "terms and conditions". The guidelines to prepare and send samples to the Core Facility are given in "Sample sheet".
García-Vílchez R, Añazco-Guenkova AM, Dietmann S, López J, Morón-Calvente V, D'Ambrosi S, Nombela P, Zamacola K, Mendizabal I, García-Longarte S, Zabala-Letona A, Astobiza I, Fernández S, Paniagua A, Miguel-López B, Marchand V, Alonso-López D, Merkel A, García-Tuñón I, Ugalde-Olano A, Loizaga-Iriarte A, Lacasa-Viscasillas I, Unda M, Azkargorta M, Elortza F, Bárcena L, Gonzalez-Lopez M, Aransay AM, Di Domenico T, Sánchez-Martín MA, De Las Rivas J, Guil S, Motorin Y, Helm M, Pandolfi PP, Carracedo A, Blanco S. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol Cancer. 2023 Jul 29;22(1):119.
 10.1186/s12943-023-01809-8 ,
10.1186/s12943-023-01809-8 ,  37516825 ,
37516825 ,  hal-04187091
hal-04187091 - Montacié C, Riondet C, Wei L, Darriere T, Weiss A, Pontvianne F, Escande ML, de Bures A, Jobet E, Barbarossa A, Carpentier MC, Aarts MGM, Attina A, Hirtz C, David A, Marchand V, Motorin Y, Curie C, Mari S, Reichheld JP, Sáez-Vásquez J. nicotianamine synthase activity affects nucleolar iron accumulation and impacts rDNA silencing and RNA methylation in Arabidopsis. J Exp Bot. 2023 May 14:erad180.
 10.1093/jxb/erad180 ,
10.1093/jxb/erad180 ,  37179467 ,
37179467 ,  HAL-04097969
HAL-04097969 - Lucas MC, Pryszcz LP, Medina R, Milenkovic I, Camacho N, Marchand V, Motorin Y, Ribas de Pouplana L, Novoa EM. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat Biotechnol. 2023 Apr 6.
 10.1038/s41587-023-01743-6 ,
10.1038/s41587-023-01743-6 ,  37024678 ,
37024678 ,  HAL-04067699
HAL-04067699 Huber LB, Kaur N, Henkel M, Marchand V, Motorin Y, Ehrenhofer-Murray AE, Marx A. A dual-purpose polymerase engineered for direct sequencing of pseudouridine and queuosine. Nucleic Acids Res. 2023 Mar 27:gkad177.
 10.1093/nar/gkad177 ,
10.1093/nar/gkad177 ,  36971106 ,
36971106 ,  HAL-04067720
HAL-04067720 - Gawade K, Plewka P, Häfner SJ, Lund AH, Marchand V, Motorin Y, Szczesniak MW, Raczynska KD. FUS regulates a subset of snoRNA expression and modulates the level of rRNA modifications. Sci Rep. 2023 Feb 20;13(1):2974.
 10.1038/s41598-023-30068-2 ,
10.1038/s41598-023-30068-2 ,  36806717 ,
36806717 ,  HAL-04015688
HAL-04015688 Pichot F, Marchand V, Helm M, Motorin Y. Data Analysis Pipeline for Detection and Quantification of Pseudouridine (ψ) in RNA by HydraPsiSeq. Methods Mol Biol. 2023;2624:207-223.
 10.1007/978-1-0716-2962-8_14 ,
10.1007/978-1-0716-2962-8_14 ,  36723818 ,
36723818 ,  HAL-03971146
HAL-03971146 - Brazane M, Dimitrova DG, Pigeon J, Paolantoni C, Ye T, Marchand V, Da Silva B, Schaefer E, Angelova MT, Stark Z, Delatycki M, Dudding-Byth T, Gecz J, Plaçais PY, Teysset L, Préat T, Piton A, Hassan BA, Roignant JY, Motorin Y, Carré C. The ribose methylation enzyme FTSJ1 has a conserved role in neuron morphology and learning performance. Life Sci Alliance. 2023 Jan 31;6(4):e202201877.
 10.26508/lsa.202201877 ,
10.26508/lsa.202201877 ,  36720500 ,
36720500 ,  HAL-03967369
HAL-03967369 - Chabronova A, van den Akker GGH, Housmans BAC, Caron MMJ, Cremers A, Surtel DAM, Wichapong K, Peffers MMJ, van Rhijn LW, Marchand V, Motorin Y, Welting TJM. Ribosomal RNA-based epitranscriptomic regulation of chondrocyte translation and proteome in osteoarthritis. Osteoarthritis Cartilage. 2023 Jan 5:S1063-4584(23)00001-8.
 10.1016/j.joca.2022.12.010 ,
10.1016/j.joca.2022.12.010 ,  36621590 ,
36621590 ,  HAL-03930713
HAL-03930713 - Pichot F, Hogg MC, Marchand V, Bourguignon V, Jirström E, Farrell C, Gibriel HA, Prehn JHM, Motorin Y, Helm M. Quantification of substoichiometric modification reveals global tsRNA hypomodification, preferences for angiogenin-mediated tRNA cleavage, and idiosyncratic epitranscriptomes of human neuronal cell-lines. Comput Struct Biotechnol J. 2022 Dec 19;21:401-417.
 10.1016/j.csbj.2022.12.020 ,
10.1016/j.csbj.2022.12.020 ,  36618980 ,
36618980 ,  HAL-03930749
HAL-03930749 Maharana S, Kretschmer S, Hunger S, Yan X, Kuster D, Traikov S, Zillinger T, Gentzel M, Elangovan S, Dasgupta P, Chappidi N, Lucas N, Maser KI, Maatz H, Rapp A, Marchand V, Chang YT, Motorin Y, Hubner N, Hartmann G, Hyman AA, Alberti S, Lee-Kirsch MA. SAMHD1 controls innate immunity by regulating condensation of immunogenic self RNA. Mol Cell. 2022 Sep 16:S1097-2765(22)00851-6.
 10.1016/j.molcel.2022.08.031 ,
10.1016/j.molcel.2022.08.031 ,  36150385 ,
36150385 ,  HAL-03787894
HAL-03787894

Formation CNRS Entreprises
( -> )
The EpiRNA-Seq Core Facility organizes the following training school (CNRS Formation Entreprises) : Epitranscriptomics: mapping and analysis of RNA modifications by Next-Generation Sequencing. This training school will be held from 17st to 19st May 2021 on the Core Facility in Nancy. For more details : https://cnrsformation.cnrs.fr/epitranscriptomics-mapping-and-analysis-of-rna-modifications-by-next-generation-sequencing?axe=140

Epitranscriptomics training school (COST Epitran)
( -> )
The NGS Core Facility in collaboration with Pr Iouri Motorine (UMR7365 IMoPA) is organizing a training school on Epitranscriptomics with the support of COST Epitran from 11-14 february 2019 in Nancy (France). This training is dedicated to young phD students or postdocs. The session in 2019 is closed. The next session will take place in February 2020, please send an email to the head of EpiRNA-Seq core facility. For more information : /sites/default/files/users/Projets/Diapositive1.png