- Facility
Epitranscriptomics & Sequencing (EpiRNA-Seq)
- Next-Generation Sequencing, RNA, RNA modifications

The Epitranscriptomics and Sequencing (EpiRNA-Seq) Core Facility is built on the expertise of the UMR7365 IMoPA in the field of RNA molecular biology to promote its main collaborative services.
Located on the Brabois-Santé campus of the University of Lorraine in Nancy, it provides to the academic and industrial scientific community high-tech resources (Illumina next generation sequencers). It is part of the European COST EPITRAN network and regularly organises training activities.
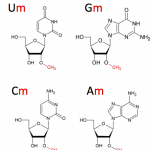
The core facility has recognized technical and methodological know-how to contribute to various projects in Epitranscriptomics (RiboMethSeq, AlkAniline-Seq, HydraPsiSeq) to address various scientific questions such as dynamics of post-transcriptional modifications in RNAs.
The EpiRNA-Seq Core Facility operates mainly in collaborative mode (involvement of members of the core facility in the steps of your sequencing project from the conception of the experimental design to the statistical analysis of the data) but the delivery mode (mainly sequencing of libraries prepared by you without involvement of the platform) is also possible under certain conditions. The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academics and industrialists).
The EpiRNA-Seq Core Facility is open to the academic and industrial scientific community at the local, national and international level.
The team is at your disposal to help you by bringing its technical and methodological know-how and to follow you from A to Z in your sequencing projects.
The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academic and industrial).
Any new collaborator must first contact the head of the core facility using the "Request form" to define the best strategy to adopt for conducting his study, and obtain a quote. Any user will agree to accept the "terms and conditions". The guidelines to prepare and send samples to the Core Facility are given in "Sample sheet".
Helm M, Schmidt-Dengler MC, Weber M, Motorin Y. General Principles for the Detection of Modified Nucleotides in RNA by Specific Reagents. Adv Biol (Weinh). 2021 Sep 17:e2100866.
 10.1002/adbi.202100866 ,
10.1002/adbi.202100866 ,  34535986 ,
34535986 ,  HAL-03349939
HAL-03349939 Marchand V, Bourguignon-Igel V, Helm M, Motorin Y. Mapping of 7-methylguanosine (m7G), 3-methylcytidine (m3C), dihydrouridine (D) and 5-hydroxycytidine (ho5C) RNA modifications by AlkAniline-Seq. Methods Enzymol. 2021;658:25-47.
 10.1016/bs.mie.2021.06.001 ,
10.1016/bs.mie.2021.06.001 ,  34517949 ,
34517949 ,  HAL-03344110
HAL-03344110 Motorin Y, Quinternet M, Rhalloussi W, Marchand V. Constitutive and variable 2'-O-methylation (Nm) in human ribosomal RNA. RNA Biol. 2021 Sep 10:1-10.
 10.1080/15476286.2021.1974750 ,
10.1080/15476286.2021.1974750 ,  34503375 ,
34503375 ,  HAL-03341858
HAL-03341858 Marchand V, Bourguignon-Igel V, Helm M, Motorin Y. Analysis of pseudouridines and other RNA modifications using hydraPsiSeq protocol. Methods. 2021 Sep 1:S1046-2023(21)00206-1.
 10.1016/j.ymeth.2021.08.008 ,
10.1016/j.ymeth.2021.08.008 ,  34481083 ,
34481083 ,  HAL-03335167
HAL-03335167 AlkAniline-Seq: A Highly Sensitive and Specific Method for Simultaneous Mapping of 7-Methyl-guanosine (m7G) and 3-Methyl-cytosine (m3C) in RNAs by High-Throughput Sequencing. Methods Mol Biol. 2021;2298:77-95.
 10.1007/978-1-0716-1374-0_5 ,
10.1007/978-1-0716-1374-0_5 ,  34085239 ,
34085239 ,  HAL-03266408
HAL-03266408 Detection and functions of viral RNA modifications: perspectives in biology and medicine. Virologie (Montrouge). 2021 Apr 1;25(2):5-20.
 10.1684/vir.2021.0884 ,
10.1684/vir.2021.0884 ,  33973850 ,
33973850 ,  HAL-03229182
HAL-03229182 Relier S, Ripoll J, Guillorit H, Amalric A, Achour C, Boissière F, Vialaret J, Attina A, Debart F, Choquet A, Macari F, Marchand V, Motorin Y, Samalin E, Vasseur JJ, Pannequin J, Aguilo F, Lopez-Crapez E, Hirtz C, Rivals E, Bastide A, David A. FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat Commun. 2021 Mar 19;12(1):1716.
 10.1038/s41467-021-21758-4 ,
10.1038/s41467-021-21758-4 ,  33741917 ,
33741917 ,  HAL-03178788
HAL-03178788

Formation CNRS Entreprises
( -> )
The EpiRNA-Seq Core Facility organizes the following training school (CNRS Formation Entreprises) : Epitranscriptomics: mapping and analysis of RNA modifications by Next-Generation Sequencing. This training school will be held from 17st to 19st May 2021 on the Core Facility in Nancy. For more details : https://cnrsformation.cnrs.fr/epitranscriptomics-mapping-and-analysis-of-rna-modifications-by-next-generation-sequencing?axe=140

Epitranscriptomics training school (COST Epitran)
( -> )
The NGS Core Facility in collaboration with Pr Iouri Motorine (UMR7365 IMoPA) is organizing a training school on Epitranscriptomics with the support of COST Epitran from 11-14 february 2019 in Nancy (France). This training is dedicated to young phD students or postdocs. The session in 2019 is closed. The next session will take place in February 2020, please send an email to the head of EpiRNA-Seq core facility. For more information : /sites/default/files/users/Projets/Diapositive1.png